Purpose and Design
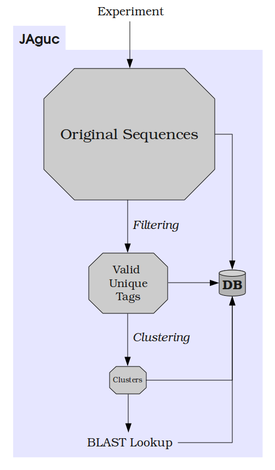
The main purpose of JAguc is to enable the analysis of biological experiments that yield hundreds of thousands of RNA sequences in a convenient and efficient way. At this time, we see two major use cases:
-
You extract some RNA from an environment sample, e.g. deep sea water. After PCR and sequencing, you get a text file with 300.000 sequences in it. You want to know what creatures lived in the original sample and how many of those. Also, you want to know if you found some now species (and killed them off while extracting, maybe?).
-
You have a set of exactly known sequences, e.g. artificially created ones. You want to test the quality of an experiment with lots of copies of those sequences.
In both cases, JAguc can be the answer. It boils down your large sequence set and checks for only a few representatives what species they belong to. This is done with filtering and clustering. Even though you get approximations there, results are still very reasonable.
Why even do that? Because databases of known sequences are awfully large and therefore searches take awfully long. JAguc minimizes the number of database searches you have to do and gives you full control over results' granularity. JAguc also keeps a record of all reductions done so you can track any result back to the original input file easily.
JAguc is not limited to the above use cases. In fact, we have even seen it being used as graphical front-end for BLAST. Application on other kinds of sequences, for example DNA or proteins, is not perfectly possible today but might not be hard to implement since most parts of JAguc should not be affected at all.
Workflow
[Details coming up soon]
Real Example
[Numbers coming up soon]
System Requirements
To run JAguc, you absolutely need a machine with
- graphical login,
- Java 6 VM,
- local installation of Mega BLAST,
- database server,
- as much RAM as possible,
JAguc itself needs about 1GB RAM for 30.000 sequences after filtering. Expect this number to increase quadratically in sequence number. Mega BLAST might need more memory, but that depends on your choice of paramters, i.e. the resulting number of clusters also. - as many and as fast CPUs as you can get and
As mentioned above, some parts can be computed in parallel while other can not. Thus, both number of cores and speed of the individual will influence runtime. - as much hard disk space as possible.
Both input files and databases can become very large. Additionally, similarity files grow quadratically in sequence number. A file for 19.000 sequences after filtering was 350MB.
We recommend you have
- Sun Java 6u10 VM or higher and
We only tested JAguc with Sun JVMs. Also, the neat Nimbus look and feel you can see on our screenshots is only available since 6u10. - a local database server installed.
JAguc writes and reads quite a bunch of data to database. If there is no network connection in between, this is much faster.
How to Get JAguc
Simple: go to our download page!
Credits
We have used the following third party libraries to make JAguc:
We thank the guys and gals that saved us a lot of work!
What about that silly name?
You are right, it is silly. We simply could not come up with a better one. It is J for Java and the other letters for the nucleotides that make up RNA. We have arranged those letters in a pronouncable way, fixing J at the first position as has become usual for software written in Java. We could not agree on one pronounciation for it, so you are free to say it as you like.


